We will work with data on the fat and protein content of items on the Burger King menu. The data are in a file named BKmenu.txt. Double-clicking on this should start a text editor (usually Notepad in Windows) with the data loaded. You will probably find it hard to work with. One strategy is to select everything and paste it into an empty Excel spreadsheet. Excel will usually separate it into columns and you can cut and paste one column at a time into the R data editor or use the scan() function. We will assume you have found some way to get the data into R. The command to get the regression equation and related information is not what you would expect. Here fat was the dependent variable and protein the independent variable. lm stands for linear model. By itself, it just returns the slope and intercept. The summary command tells it to return the summary table below.
> summary(lm(formula = fat ~ protein))
Call:
lm(formula = fat ~ protein)
Residuals:
Min 1Q Median 3Q Max
-11.726 -8.772 1.239 7.029 20.052
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 6.4113 2.6466 2.423 0.0217 *
protein 0.9769 0.1212 8.057 5.4e-09 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 9.311 on 30 degrees of freedom
Multiple R-Squared: 0.6839, Adjusted R-squared: 0.6734
F-statistic: 64.92 on 1 and 30 DF, p-value: 5.402e-09
There is much more here than you really need so we have put the basic information in red. You can read off the regression equation as fat = 6.4113 + 0.9769*protein. R2 = 0.6839 = 68.39% and se = 9.311. The bad news is that fat tends to go up when protein content goes up (positive slope). The good news is that there is lots of scatter (R2 = 68.39% ) and so you may find exceptions.
You can also compute or graph the residuals.
> residuals(lm(formula = fat ~ protein))
1 2 3 4 5 6
4.2599876 7.3757304 3.6998103 6.8155532 -9.9946466 -7.9483495
7 8 9 10 11 12
-10.6937153 -6.6011210 -6.5316753 -10.0872409 8.1442448 -10.7168638
13 14 15 16 17 18
6.9127591 -8.1566866 7.6812735 5.6812735 -0.2492808 -2.1335380
19 20 21 22 23 24
5.6349763 11.0285020 2.7275706 20.0516505 15.8896106 7.7275706
25 26 27 28 29 30
10.7275706 6.6349763 -10.1335380 -11.1103894 -10.1335380 -6.4113208
31 32
-8.3650237 -11.7261323
> plot(protein,residuals(lm(formula = fat ~ protein)))
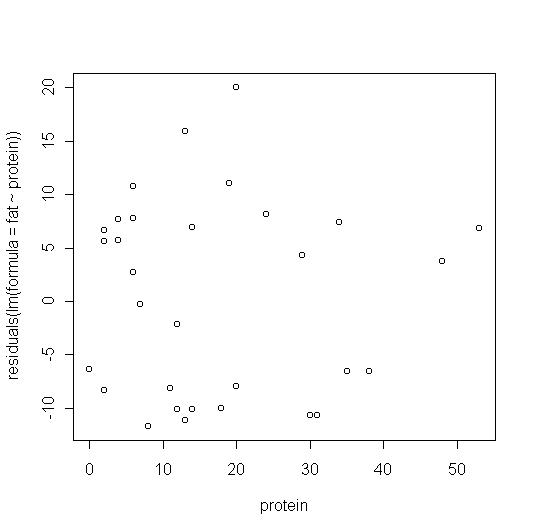
These look reasonably random but not clumped around zero. Instead there seems to be a group of residuals around 10 and another around -10.
If you plan to do much with the residuals, you may wish to store them in a variable for further work. For example, here they are stored in a variable res and then a histogram is made.
> res = residuals(lm(formula = fat ~ protein)) > hist(res)
The histogram is not reproduced here but it shows signs of the bimodality mentioned above.
© 2006 Robert W. Hayden